|
I am a research assistant at the University of California, Riverside (UCR), where we leverage computational algorithms to design compact Cas9-sgRNA libraries for CRISPR genome editing across fungal species. This work facilitates the production of novel compounds, unlocking fungi's potential for diverse applications, including sustainable biofuels, eco-friendly biomaterials, life-saving pharmaceuticals, and innovative food products. I earned my MS in Chemical and Environmental Engineering from University of California, Riverside under supervision of Professor Ian Wheeldon , and prior to that, I completed my bachelor’s degree in chemical engineering at the University of Tehran. Throughout my research journey, I had the privilege of collaborating closely with Professor Stefano Lonardi from the University of California, Riverside, and Professor Abbas Ali Khodadadi from the University of Tehran, whose mentorship greatly enriched my work. Email / Google Scholar / GitHub / LinkedIn / CV |

|
|
|
My primary research centers on computational biology and CRISPR-Cas9 knockout screenings. In the past, I have explored diverse areas including drug delivery, metal-organic frameworks (MOFs), and molecular dynamics simulations. |
|
Amirsadra Mohseni*, Reyhane Ghorbani Nia*, Aida Tafrishi, Xinzhan Liu, Jason Stajich, Stefano Lonardi, Ian Wheeldon In this work, we introduce ALLEGRO, a computational method leveraging combinatorial optimization to design minimal gRNA libraries for thousands of fungal species. ALLEGRO efficiently identifies orthologous genes, extracts candidate gRNAs, predicts their activity scores, and employs linear programming to generate the smallest high-activity gRNA libraries within minutes, enabling precise and scalable genome editing. |
|
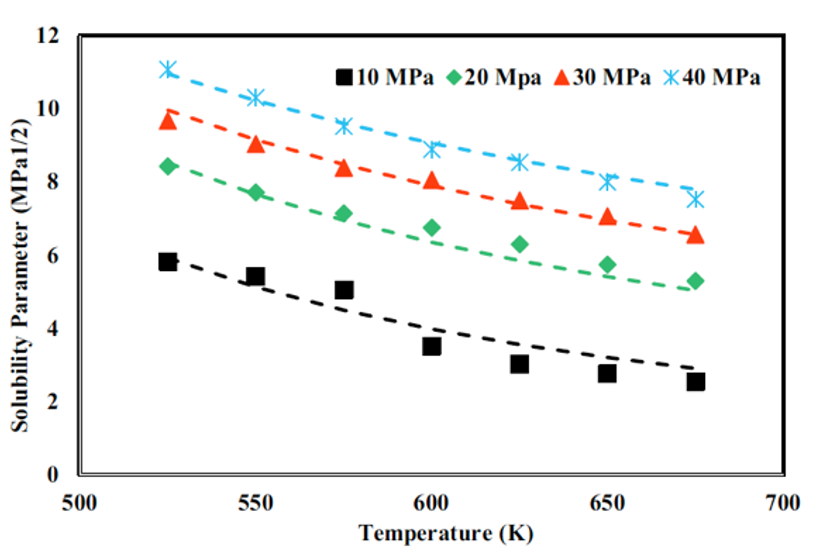
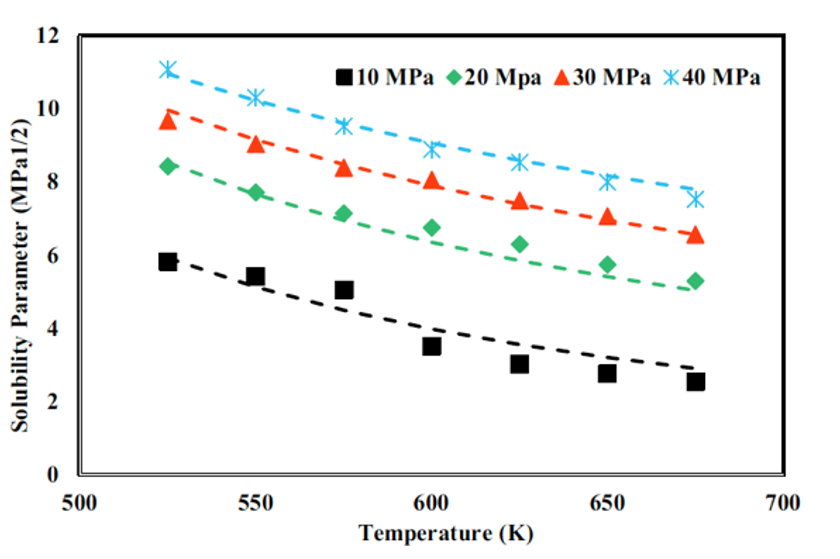
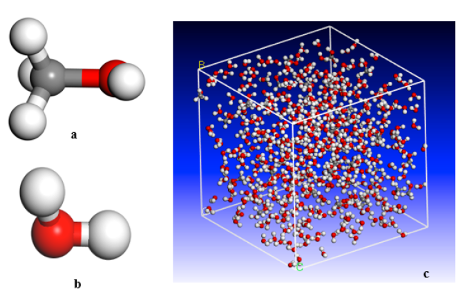
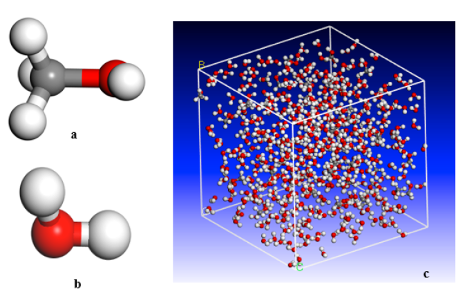
Hojatollah Moradi, Dr. Hedayat Azizpour, Dr. Parissa Khadiv-Parsi, Reyhane Ghorbani Nia Journal: Chemical Engineering Technology, Wiley, 2023 In this study, we explored the solubility parameters of supercritical methanol and ethanol through molecular dynamics simulations across varying temperatures and pressures. The predicted solubility parameters closely align with theoretical data, validating our approach. Electrostatic interactions were modeled using the Ewald summation method, while van der Waals interactions were handled with the atom-based summation method. Additionally, our findings demonstrate that increasing density linearly improves solubility. |
|
Hojatollah Moradi, Dr. Hedayat Azizpour, Dr. Kamran Keynejad, Reyhane Ghorbani Nia, Dr. Amin Esmaeili Journal: Chemical Engineering Technology, Wiley, 2023 In this work, we utilize molecular dynamics (MD) simulation and empirical data to predict the density of water-methanol mixtures at 293.15–303.15K and atmospheric pressure. Our approach employs the Ewald summation method for electrostatic interactions and the atom-based method for van der Waals interactions. We further apply the Jouyban-Acree (J-A) model and density correlation methods for two-component mixtures to analyze MD simulation results across various temperatures and methanol mole fractions. |
|
|
|
Last update: December, 2024 |